Pre-processing¶
Convert CI data to NWB file format
Data to NWB¶
-
cicada.preprocessing.cicada_data_to_nwb.convert_data_to_nwb(data_to_convert_dir, default_convert_to_nwb_yml_file, nwb_files_dir)[source]¶ Convert all default_config_data_for_conversion located in dir_path and put it in NWB format then create the file. Use the yaml file contains in dir_path to convert the default_config_data_for_conversion. A yaml file with in its name session_data and one with subject_data must be in directory. Otherwise nothing will happend. A yaml file with abf in its name will need to be present to convert the abf default_config_data_for_conversion.
Parameters:
-
cicada.preprocessing.cicada_data_to_nwb.create_convert_class(class_name, config_dict, converter_dict, nwb_file, yaml_path, files, dir_path)[source]¶ Parameters: - class_name –
- config_dict –
- converter_dict –
- nwb_file –
- yaml_path –
- files –
- dir_path –
Returns:
-
cicada.preprocessing.cicada_data_to_nwb.create_nwb_file(subject_data_yaml_file, session_data_yaml_file)[source]¶ Create an NWB file object using all metadata containing in YAML file
Parameters:
-
cicada.preprocessing.cicada_data_to_nwb.filter_list_according_to_keywords(list_to_filter, keywords, keywords_to_exclude)[source]¶ Conditional loop to remove all files or directories not containing the keywords # or containing excluded keywords. Inplace list modification
Parameters: - Exemples:
>>> print(filter_list_of_files(["file1.py", "file2.c", "file2.h"],"2","h")) ["file2.c"]
-
cicada.preprocessing.cicada_data_to_nwb.filter_list_of_files(dir_path, files, extensions, directory=None)[source]¶ Take a list of file names and either no extensions (empty list or None) and remove the directory that starts by “.” or a list of extension and remove the files that are not with this extension. It returns a new list
Parameters: - Exemples:
>>> print(filter_list_of_files(["file1.py", "file2.c", "file3.h"],"py")) ["file1.py"]
Run preprocessing¶
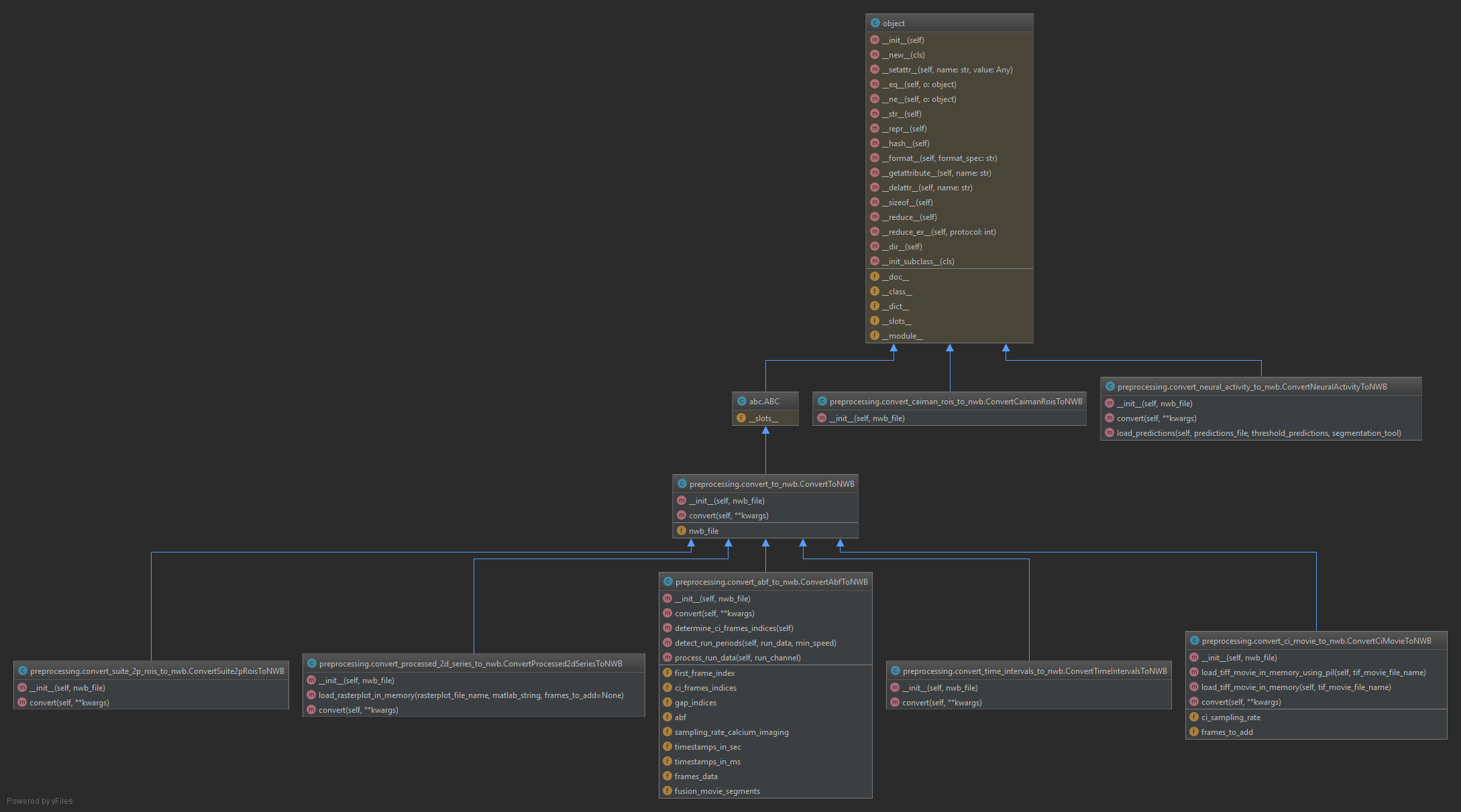
NWB file class¶
Suite 2P ROIs¶
2D series¶
-
class
cicada.preprocessing.convert_processed_2d_series_to_nwb.ConvertProcessed2dSeriesToNWB(nwb_file)[source]¶ Class to convert 2D series to NWB
-
convert(**kwargs)[source]¶ Convert the data and add to the nwb_file
Parameters: **kwargs – arbitrary arguments
-
Calcium imaging movie¶
ABF¶
-
class
cicada.preprocessing.convert_abf_to_nwb.ConvertAbfToNWB(nwb_file)[source]¶ Class to convert ABF data to NWB
-
convert(**kwargs)[source]¶ The goal of this function is to extract from an Axon Binary Format (ABF) file its content and make it accessible through the NWB file. The content can be: LFP signal, piezzo signal, speed of the animal on the treadmill. All, None or a few of these informations could be available. One information always present is the timestamps, at the abf sampling_rate, of the frames acquired by the microscope to create the calcium imaging movie. Such movie could be the concatenation of a few movies, such is the case if the movie need to be saved every x frames for memory issue for ex. If the movie is the concatenation of many, then there is an option to choose to extract the information as if 2 frames concatenate are contiguous in times (such as then LFP signal or piezzo would be match movie), or to add interval_times indicating at which time the recording is on pause and at which time it’s starting again. The time interval containing this information is named “ci_recording_on_pause” and you can get it doing: if ‘ci_recording_on_pause’ in nwb_file.intervals: pause_intervals = nwb_file.intervals[‘ci_recording_on_pause’]
Parameters: - **kwargs (dict) – kwargs is a dictionary, potentials keys and values types are:
- abf_yaml_file_name – mandatory parameter. The value is a string representing the path
- file_name of the yaml file associated to this abf file. In the abf (and) –
- frames_channel – mandatory parameter. The value is an int representing the channel
- the abf in which is the frames timestamps data. (of) –
- abf_file_name – mandatory parameter. The value is a string representing the path
- file_name of the abf file. (and) –
-
detect_run_periods(run_data, min_speed)[source]¶ Using the data from the abf regarding the speed of the animal on the treadmill, return the speed in cm/s at each timestamps as well as period when the animal is moving (using min_speed threshold)
Parameters: Returns: List of movements periods speed_during_movement_periods (list) : List of subject speed during movements speed_by_time (list) : List of subject speed by time
Return type: mvt_periods (list)
-
determine_ci_frames_indices()[source]¶ Using the frames data channel, estimate the timestamps of each frame of the calcium imaging movie. If there are breaks between each recording (the movie being a concatenation of different movies), then there is an option to either skip those non registered frames that will be skept in all other data (lfp, piezzo, …) or to determine how many frames to add in the movie and where so it matches the other data recording in the abf file
-
Utils¶
-
class
cicada.preprocessing.utils.ComparableItem(value)[source]¶ Make it possible to sort a list of items of different types, such as int and string
-
cicada.preprocessing.utils.class_name_to_file_name(class_name)[source]¶ Transform the string representing a class_name, by removing the upper case letters, and inserting before them an underscore if 2 upper case letters don’t follow. Underscore are also inserted before numbers ex: ConvertAbfToNWB -> convert_abf_to_nwb :param class_name: string :return:
-
cicada.preprocessing.utils.flatten(list)[source]¶ Flatten a nested list no matter the nesting level
Parameters: list (list) – List to flatten Returns: List without nest Examples
>>> flatten([1,2,[[3,4],5],[7]]) [1,2,3,4,5,7]
-
cicada.preprocessing.utils.get_continous_time_periods(binary_array)[source]¶ take a binary array and return a list of tuples representing the first and last position(included) of continuous positive period :param binary_array: :return:
-
cicada.preprocessing.utils.load_tiff_movie_in_memory(tif_movie_file_name, frames_to_add=None)[source]¶ Load tiff movie from filename using Scan Image Tiff
Parameters: tif_movie_file_name (str) – Absolute path to tiff movie Returns: Tiff movie as 3D-array Return type: tiff_movie (array)
-
cicada.preprocessing.utils.load_tiff_movie_in_memory_using_pil(tif_movie_file_name, frames_to_add=None)[source]¶ Load tiff movie from filename using PIL library
Parameters: - tif_movie_file_name (str) – Absolute path to tiff movie
- frames_to_add – dict with key an int representing the frame index after which add frames. the value is the number of frames to add (integer)
Returns: Tiff movie as 3D-array
Return type: tiff_movie (array)
-
cicada.preprocessing.utils.merging_time_periods(time_periods, min_time_between_periods)[source]¶ Take a list of pair of values representing intervals (periods) and a merging thresholdd represented by min_time_between_periods. If the time between 2 periods are under this threshold, then we merge those periods. It returns a new list of periods. :param time_periods: list of list of 2 integers or floats. The second value represent the end of the period, the value being included in the period. :param min_time_between_periods: a float or integer value :return: a list of pair of list.
-
cicada.preprocessing.utils.sort_by_param(nwb_path_list, param_list)[source]¶ Sort NWB files depending on a list of parameters
Parameters: Returns: List of NWB files sorted
Return type: nwb_sorted_list (list)
-
cicada.preprocessing.utils.update_frames_to_add(frames_to_add, nwb_file, ci_sampling_rate)[source]¶ Update frames_to_add (dict), based on pause_intervals and ci_frames_time_series :param frames_to_add: dict, with key an int representing the frame index after which add frames. :param the value is the number of frames to add: :type the value is the number of frames to add: integer :param nwb_file: nwb file, will get nwb_file.intervals[‘ci_recording_on_pause’] and :param nwb_file.get_acquisition: :type nwb_file.get_acquisition: “ci_frames”
Returns:
Appendix¶